research articles
Sweet et al.
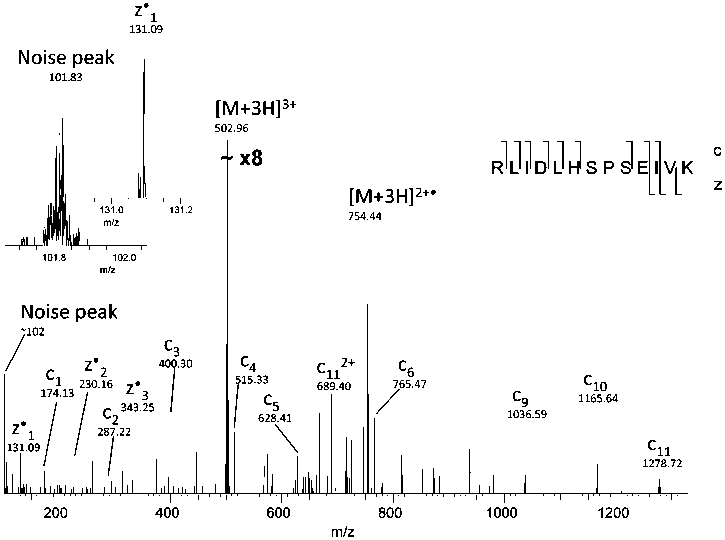
Figure 1. Noise peak at m/z ~102. Insets show enlargements of the noise peak and a true fragment peak of similar m/z.
Table 3. Effect of Presearch Trimming of ECD DTA files on
the Number of Mascot Identificationsa
incorrect precursor determination. Precursor mass tolerances
of 2.1 and 3.1 Da do not increase the number of identifications,
indicating that the Bioworks-generated DTA contains the
correct monoisotopic mass. When the second or third isotopic
peak was selected for fragmentation, the mass in the DTA file
generated by Bioworks was automatically corrected to the
monoisotopic value. (As a result of the high precursor threshold
for fragmentation, the true monoisotopic ion was always clearly
detected.)
Preprocessing of ECD Spectra. Electrical Noise Peak. The
results of the initial searches, as reported in Tables 1 and 2,
indicate an identification success rate of approximately 70%
for CID mass spectra and 45% for ECD mass spectra. We
examined the ECD spectra for generic features which may be
detrimental to the identification success rate. Electrical noise
peaks are unfortunate but commonplace features of mass
spectra. In all FT-ICR data from our instrument, we observe a
peak of this kind at m/z 102. No equivalent peaks are observed
in the LTQ linear ion trap mass spectra. The peak is broad and
lacks isotope peaks, easily distinguishing it from true analyte
peaks (Figure 1). Removing the region in which this peak occurs
(101.7-102.1 m/z) from the DTA files is a straightforward way
to improve the ratio of informative to uninformative peaks for
the database search. This increases the Mascot and OMSSA
identification scores and therefore number of identifications
(Tables 3 and 4, row 2). The noise peak removal could in theory
also remove a z-dot ion from C-terminal valine; however, this
will have no effect in practice, since a vast majority of tryptic
peptides have lysine or arginine at their C-terminus. (The noise
peak would also obscure the valine z-dot ion.)
Coeluting Peaks of Similar m/z (within the Precursor
Isolation Window). To maximize the transfer of large numbers
of precursor ions from the linear ion trap into the ICR cell for
ECD fragmentation, a precursor isolation window larger than
that selected for LTQ CID is usually employed.12 This window
typically ranges from 3 to 10 m/z.5,13 Here, we use an isolation
width of 6 m/z. During analysis of complex proteomic samples,
5478 Journal of Proteome Research • Vol. 8, No. 12, 2009
forward reverse
presearch hits (2+) hits ID rate
ECD Search (3341 DTAs) Mascot
|
No DTA trimming |
1468 (870) |
7 |
43.9 |
|
Remove noise peak |
1509 (898) |
12 |
45.2 |
|
Remove precursor window |
1556 (934) |
9 |
46.6 |
|
Remove precursor window. |
1586 (964) |
13 |
47.5 |
|
Remove noise peak |
1609 (972) |
10 |
48.2 |
|
Remove noise peak |
1643 (1006) |
16 |
49.2 |
and precursor window.
Remove all nonfragments
(MH to MH - 140 m/z).
a Identifications from doubly-charged precursors shown in parentheses.
coelution of peptides of similar m/z is frequently observed.11
In contrast to the LTQ CID process, peaks in the isolation
window are not fragmented to completion during ECD making
any coelution of peptides more readily apparent. Both OMSSA
and Mascot have options to remove the intact precursor peak
(these options are employed in all the searches described here);
however, this only removes the precursor and its isotopes
according to the user-defined fragment ion mass tolerance, and
not any coeluting peptide or noise peaks within the precursor
isolation window. We investigated whether removing the
complete isolation window from the DTA files prior to database
searching would improve the resulting identifications (Tables
3 and 4). An example of an additional identification resulting
from the removal of the isolation window is shown in Figure
2. It should be noted that bona fide fragment peaks may
inadvertently be removed during this trimming process. We
remove the 6 m/z region around the precursor. For a typical
1500 Da, 2+ tryptic peptide, the acquired MS/MS ECD mass
spectrum ranges from 100 to 1500 m/z. Therefore, we remove
More intriguing information
1. Behavioural Characteristics and Financial Distress2. Does adult education at upper secondary level influence annual wage earnings?
3. Permanent and Transitory Policy Shocks in an Empirical Macro Model with Asymmetric Information
4. Developmental changes in the theta response system: a single sweep analysis
5. BEN CHOI & YANBING CHEN
6. NVESTIGATING LEXICAL ACQUISITION PATTERNS: CONTEXT AND COGNITION
7. The Values and Character Dispositions of 14-16 Year Olds in the Hodge Hill Constituency
8. Word searches: on the use of verbal and non-verbal resources during classroom talk
9. Picture recognition in animals and humans
10. The name is absent