Database Search Strategies for Proteomic Data Sets
research articles
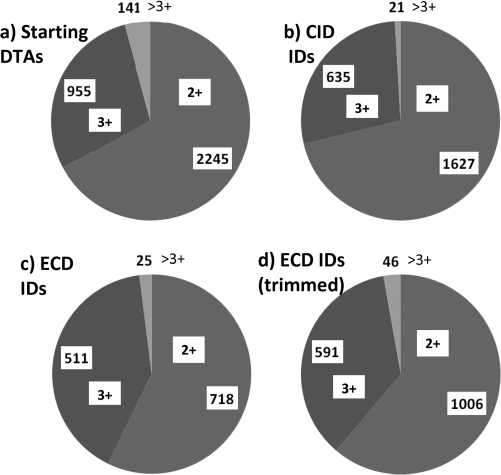
Figure 4. DTAs and identifications by charge-state. (a) Data set
of 3341 DTAs (CID and ECD pairs); (b) CID identifications (2283)
using Mascot with subsequent filtering; (c) ECD identifications
(1254) using Mascot with subsequent score filtering; (d) ECD
identifications (1643) using trimmed DTAs, Mascot and subse-
quent score and mass accuracy filtering.
lower intensity than z fragments (with the exception of y
fragments N-terminal to proline); runs of consecutive c or z
■ Conflicts within group ι≡ Reverse hits within peptide score boundaries
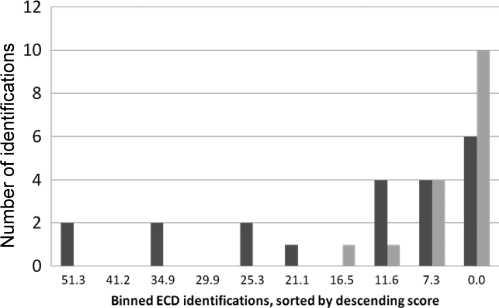
Figure 6. Distribution of identification conflicts and reverse hits
by ECD identification Mascot peptide score. The abscissa shows
1560 ECD identifications, with paired CID identifications, binned
according to descending Mascot score: ten bins, each containing
156 ECD IDs, labeled with lowest score in bin.
fragments. The manual validation suggests that the number
of false-positives within this subset of 83 identifications is
greater than for the data set overall, with four putative false
positives giving a FDR of 5%. Nonetheless, 95% of these ECD
identifications pass manual scrutiny. Two examples of low-
scoring ECD identifications are shown in Figure 7.
Comparison with Existing ETD Spectral Processing Algo-
rithm. In recent work by Good et al., an algorithm designed
for ETD fragmentation mass spectra preprocessing was re-
removing neutral Io
precursor {2+ DTAs)
DTA #
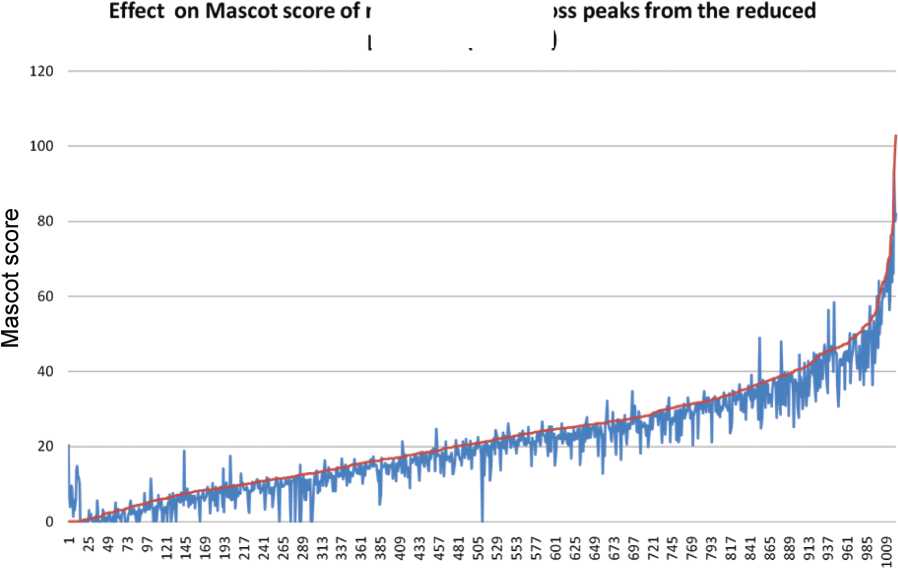
Figure 5. Effect on Mascot score of removing non-c, z, y fragment peaks within 140 m/z of the charge-reduced precursor (RP-140) of
doubly charged peptides. A total of 3341 ECD DTAs were searched, without neutral loss peak removal and with neutral loss peak
removal resulting in 972 and 1006 2+ identifications, respectively. A total of 1021 DTAs resulted in an identification in one or both
searches. The identifications are plotted, by ascending Mascot score of RP-140 trimmed version (red). The identification score for each
DTA prior to RP-140 trimming is shown alongside (blue). In both cases, the precursor window and noise peak at m/z 102 were removed.
Reverse hits or rejected hits (unacceptably large ppm error) were assigned a score of zero.
Journal of Proteome Research • Vol. 8, No. 12, 2009 5481
More intriguing information
1. News Not Noise: Socially Aware Information Filtering2. The Role of Trait Emotional Intelligence (El) in the Workplace.
3. AN EMPIRICAL INVESTIGATION OF THE PRODUCTION EFFECTS OF ADOPTING GM SEED TECHNOLOGY: THE CASE OF FARMERS IN ARGENTINA
4. Conservation Payments, Liquidity Constraints and Off-Farm Labor: Impact of the Grain for Green Program on Rural Households in China
5. The name is absent
6. Opciones de política económica en el Perú 2011-2015
7. Co-ordinating European sectoral policies against the background of European Spatial Development
8. sycnoιogιcaι spaces
9. The name is absent
10. The name is absent