research articles
Sweet et al.
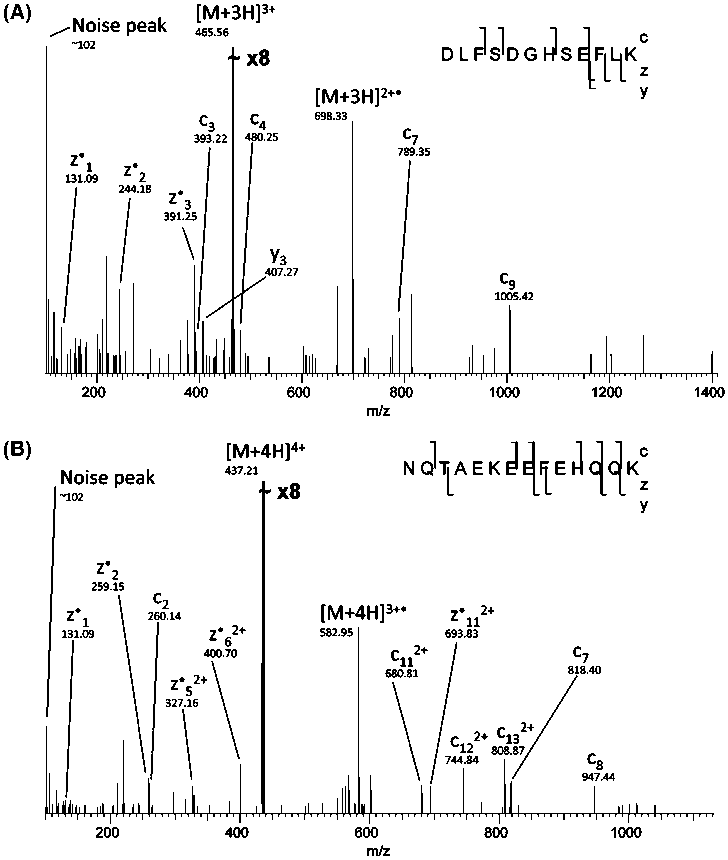
Figure 7. (A) ECD identification with a Mascot peptide score of 0.36. (B) ECD identification with a Mascot peptide score of 1.96. In both
cases, all fragments are identified with mass accuracy better than 10 ppm. Mass spectra are shown as acquired, i.e., prior to trimming.
ported.9 ETD and ECD result in similar fragmentation patterns
and it is therefore likely that the described ETD algorithm
would be useful for ECD spectral processing. We compared the
two data processing methods directly, using the same set of
DTAs (generated by the Good algorithm from raw files). The
results are shown in Table 5. The ETD algorithm allows removal
of the precursor window, neutral losses up to 60 Da from the
charge-reduced precursor and the charge-reduced precursor
ions themselves. Both processing options result in an increase
in identifications compared to the unprocessed DTAs, with the
ECD processing algorithm resulting in the most identifications.
These searches resulted in fewer identifications than the
equivalent searches using DTAs generated by Bioworks. Bio-
works automatically corrects the precursor mass to the monoiso-
topic value even if the second isotopic peak was selected for
fragmentation. The DTAs generated using the ETD algorithm
may be out by 1 Da if the second isotopic peak was selected
for fragmentation. While these identifications are recovered by
filtering two windows, one around 0 Da error and the second
around 1 Da error, the likelihood of a false-positive is now
increased.
|
Table 5. Comparison with Good9 Algorithma | |||
|
presearch |
forward |
reverse hits I |
D rate |
|
ECD search (3341 DTAs); Good Algorithm; Mascot | |||
|
DTAs generated using |
1423 (848) |
14 |
42.6 |
|
DTAs generated and processed |
1470 (873) |
14 |
44.0 |
|
DTAs generated using Good |
1507 (915) |
15 |
45.1 |
a Identifications from doubly-charged precursors are shown in
parentheses. To achieve the estimated FDR of 1%, results were filtered
according to Mascot scores (scores of 4.09, 0.79 and 8.4, respectively).
Applicability to Phosphoproteomic Data Set. In earlier
work, we identified over 900 phosphopeptides from a similar
mouse whole-cell lysate sample after TiO2-based phosphopep-
tide enrichment.5 From a total of 6080 ECD DTAs, 1087
phosphopeptides identifications were made (with redundancy).
These identifications were made using the same search (OMSSA)
and postsearch strategies (filtering by precursor mass error and
5482 Journal of Proteome Research • Vol. 8, No. 12, 2009
More intriguing information
1. INTERACTION EFFECTS OF PROMOTION, RESEARCH, AND PRICE SUPPORT PROGRAMS FOR U.S. COTTON2. Research Design, as Independent of Methods
3. How we might be able to understand the brain
4. The name is absent
5. Language discrimination by human newborns and by cotton-top tamarin monkeys
6. Life is an Adventure! An agent-based reconciliation of narrative and scientific worldviews
7. Literary criticism as such can perhaps be called the art of rereading.
8. Do Decision Makers' Debt-risk Attitudes Affect the Agency Costs of Debt?
9. Gerontocracy in Motion? – European Cross-Country Evidence on the Labor Market Consequences of Population Ageing
10. Valuing Access to our Public Lands: A Unique Public Good Pricing Experiment