Table 2: Transfer of antibiotic resistance from donor to recipient strains and transfer frequencies
|
Set No. |
Donor strain |
Recipient strain |
Transconjugant |
Conjugation |
|
I |
E. coliEC3 (ACCoTNxCp) |
S. typhi B72 (Sensitive) |
S. typhi B72 [pEC3] (ACCoT) |
0.39 × 10-7 |
|
S. typhi B72 [pEC3] (ACCoT) |
E. coli C600 (Nx) |
E. coli C600 [pEC3] (ACCoT Nx) |
0.98 × 10-5 | |
|
II |
K. pneumoniae K1 (ACCoT) |
S. typhi B72 (Sensitive) |
S. typhi B72 [pK1] (ACCoT) |
0.25 × 10-7 |
|
S. typhi B72 [pK1] (ACCoT) |
E. coli C600 (Nx) |
E. coli C600 [pK1] (ACCoT Nx) |
0.92 × 10-6 | |
|
III |
P. vulgaris Prv2 (ACCoT) |
E. coli C600 (Nx) |
E. coli C600 [pPrv2] (ACCoT Nx) |
0.98 × 10-6 |
|
E. coli C600 [pPrv2] (ACCoT Nx) |
S. typhi B72 (Sensitive) |
S. typhi B72 [pPrv2] (ACCoT) |
0.12 × 10-7 |
A=ampicillin, C=chloramphenicol, Co=Cotrimoxazole, T=tetracycline, Nx=nalidixic acid, Cp=Ciprofloxacin
The MDR E. coli showing resistance to A, C, Co, T,
Nx, and ciprofloxacin (Cp) and K. pneumoniae
(resistant to ACCoT) transferred ACCoT-resistance to
the antibiotic sensitive S. typhi; NxCp-resistance was
not transferred, and the transfer frequencies were 0.39
× 10-7 and 0.25 × 10-7, respectively. In the secondary
transfer studies, all types of transconjugants obtained
from the primary conjugation studies were used as
the donors that transferred ACCoT-resistance to E.
coli C600 with transfer frequencies 0.98 × 10-5 and 0.92
× 10-6, respectively. The donor P. vulgaris strain Prv2
transferred the complete resistance pattern of ACCoT
to the antibiotic sensitive S. typhi strain through the
primary recipient E. coli C600 with transfer
frequencies 0.98 × 10-6 and 0.12 × 10-7, respectively.
Plasmid profile
The MDR S. typhi isolates (resistance pattern ACCoT)
obtained during 1991 enteric fever outbreak in several
parts of West Bengal were screened for the presence
of plasmid. The all S. typhi strains from the three
different epidemic zones of West Bengal contained
plasmids, which co-migrated with each other. Fig. 1
shows the plasmids of three different S. typhi strains
BS13, AS12, and M54 collected from Bagnan, Asansol
and Khardah, respectively.
Figure 1: Agarose gel electrophoresis of plasmid
DNAs from sporadic isolates and outbreak causing
isolates of Salmonella typhi
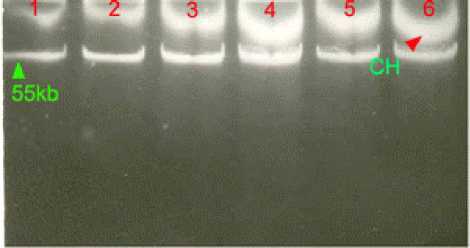
Lane 1: S. typhi strain AS12 (ACCoT) of 1991; Lane 2: S. typhi
strain BS13 (ACCoT) of 1991; Lane 3: S. typhi strain M54
(ACCoT) of 1991; Lane 4: S. typhi strain B2/92 (ACCoT) of
1992; Lane 5: E. coli C600 transconjugant (pB2/92); Lane 6: S.
typhi strain D1/01 (NxACCoT) of the year 2000. CH,
chromosome.
Recurrence of same resistance pattern (ACCoT) was
noticed in S. typhi strains during 1992 and 2000 too.
These strains contained plasmids, which co-migrated
with plasmid DNA obtained from S. typhi isolates of
1991.
Figure 2: Agarose gel electrophoresis of the plasmid
DNAs isolated from Salmonella typhi and the
transconjugants
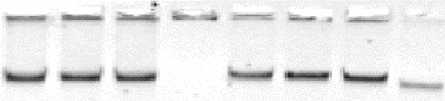
1 2 3 4 5 6 7 8
Lane 1: S. typhi (ACCoT) of 1991(BS13); Lane 2: E. coli C600
primary transconjugant (pSTBS13); Lane 3: S. typhi B72
secondary transconjugant (pSTBS13); Lane 4: S. typhi B72
(sensitive to antibiotics), Lane 5: S. typhi (NxACCoT) of 2000
(BS225); Lane 6: E. coli C600 primary transconjugant
(pSTBS225); Lane 7: S. typhi B72 secondary transconjugant
(pSTBS225); Lane 8: plasmid size marker of 53.7 kb from E.
coli V517.
Fig. 2 illustrates that the Plasmid DNAs isolated from
the primary and secondary transconjugants co-
migrated with the plasmid isolated from their
corresponding donor strains, and are about 55 kb. The
antibiotic sensitive strains of S. typhi did not show any
plasmid band in the gel.
In search of the plasmid conferring multi drug
resistance to A, C, Co, and T among MDR S. typhi
isolates, we isolated plasmid DNA from MDR E. coli,
K. pneumoniae and P. vulgaris, which showed ACCoT
resistance pattern. The strains, E. coli, K. pneumoniae,
and P. vulgaris as well as their transconjugants
showed plasmid band co-migrated with the plasmid
More intriguing information
1. Macroeconomic Interdependence in a Two-Country DSGE Model under Diverging Interest-Rate Rules2. An alternative way to model merit good arguments
3. CGE modelling of the resources boom in Indonesia and Australia using TERM
4. Mergers under endogenous minimum quality standard: a note
5. American trade policy towards Sub Saharan Africa –- a meta analysis of AGOA
6. The name is absent
7. PERFORMANCE PREMISES FOR HUMAN RESOURCES FROM PUBLIC HEALTH ORGANIZATIONS IN ROMANIA
8. The name is absent
9. From Aurora Borealis to Carpathians. Searching the Road to Regional and Rural Development
10. The Structure Performance Hypothesis and The Efficient Structure Performance Hypothesis-Revisited: The Case of Agribusiness Commodity and Food Products Truck Carriers in the South