1106 K. Hollands, D. J. Lee, G. S. Lloyd and S. J. W. Busby ■
CRP
-10
-35
aer

TGTGA
TCACA
TaaaGTTT
Gccgataa
promoter fragment
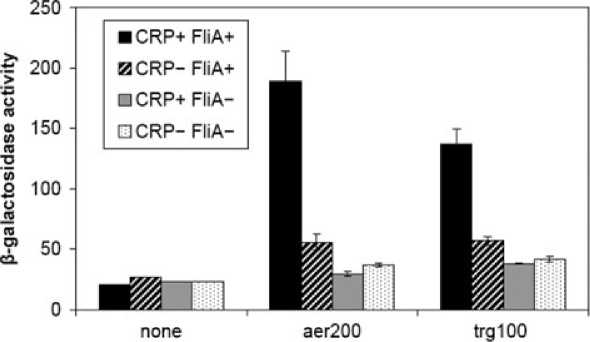
Fig. 7. Activation of the s28-dependent trg promoter by CRP.
A. Sequence alignment of the aer and trg regulatory regions. The -10 and -35 elements of the s28-dependent aer and trg promoters, and the
DNA sites for CRP, are highlighted. The consensus sequences for CRP and Es28 binding (Busby and Ebright, 1999; Koo et al., 2009) are
shown below the alignment. Asterisks below the sequence specify bases that are identical in the two sequences.
B. Effect of CRP on expression from the trg promoter. The figure shows b-galactosidase activities (in Miller units) measured in strain M182
DfliA containing pKXH100 (CRP+ FliA+), strain M182 DfliA Dcrp containing pKXH100 (CRP- FliA+), strain M182 DfliA containing ‘empty’ pET21a
(CRP+ FliA-) or strain M182 DfliA Dcrp containing ‘empty’ pET21a (CRP- FliA-), each carrying the trg100::IacZfusion cloned in pRW50. pRW50
carrying the pUC9 linker was included as a negative control (‘none’), and the aer200::lacZ fusion, cloned in pRW50, was included as a
positive control. Data shown are averages from three independent experiments, and error bars indicate one standard deviation.
one helical turn upstream, CRP can activate s38-, but not
s70-dependent transcription.
Activation at the aer promoter requires AR1 of CRP that
likely contacts aCTD (Hollands et al., 2007). Our results
show that the two aCTDs of Es28 contact DNA both
upstream and downstream of CRP, although note that we
cannot prove that both contacts occur simultaneously.
The finding that one aCTD binds downstream of CRP at
the aer promoter was surprising. Structural modelling of
the CRP-RNA polymerase-DNA complex at a Class I pro-
moter, where the DNA site for CRP is centred at position
-61.5, indicates that one aCTD is tightly sandwiched
between CRP and s70, such that it can simultaneously
contact DNA, AR1 on CRP and s70 domain 4 (Chen et al.,
2003; Lawson et al., 2004). As the DNA site for CRP at the
aer promoter is located 12 bp downstream, it appears that
there cannot be sufficient space for aCTD to fit between
the CRP dimer and the promoter-bound sigma factor. We
modelled the structure of the CRP-RNA polymerase-DNA
complex at the aer promoter by combining the crystal
structure of the CRP-aCTD-DNA complex (Benoff et al.,
2002) with the EsA.-fork junction DNA structure (Murakami
et al., 2002) and, as expected, we found that there is a
clash between the predicted locations of the aCTD down-
stream of CRP and domain 4 of s, which contacts
the promoter -35 element (K. Hollands and D.J. Lee,
unpublished). This leads us to propose a model in which
the organization of the CRP-Es28-DNA complex at the aer
promoter differs from that of the CRP-Es70-DNA complex
at a Class I s70-dependent promoter (Fig. 8A). Our
FeBABE footprinting data indicate that the juxtaposition of
CRP and the downstream aCTD at the aer promoter is the
same as at the Class I promoter. This implies that it must
be domain 4 of s28 that is positioned differently within the
CRP-Es28-DNA complex, compared with domain 4 of s70
within the CRP-Es70-DNA complex at a Class I promoter.
This is supported by the observation that the -10 and -35
elements at s28-dependent promoters are located 2-3 bp
closer together than at promoters served by Es70
(Fig. 8B), suggesting that the contact site for s28 domain 4
on promoter DNA may lie several bases downstream of
that for s70 domain 4 at a s70-dependent promoter. The
© 2009 The Authors
Journal compilation © 2009 Blackwell Publishing Ltd, Molecular Microbiology, 75, 1098-1111
More intriguing information
1. The Social Context as a Determinant of Teacher Motivational Strategies in Physical Education2. The economic value of food labels: A lab experiment on safer infant milk formula
3. The name is absent
4. FUTURE TRADE RESEARCH AREAS THAT MATTER TO DEVELOPING COUNTRY POLICYMAKERS
5. EXECUTIVE SUMMARY
6. The name is absent
7. The name is absent
8. Improvements in medical care and technology and reductions in traffic-related fatalities in Great Britain
9. Iconic memory or icon?
10. Does Presenting Patients’ BMI Increase Documentation of Obesity?