1104 K. Hollands,D. J. Lee,G. S. LloydandS. J. W. Busby ■
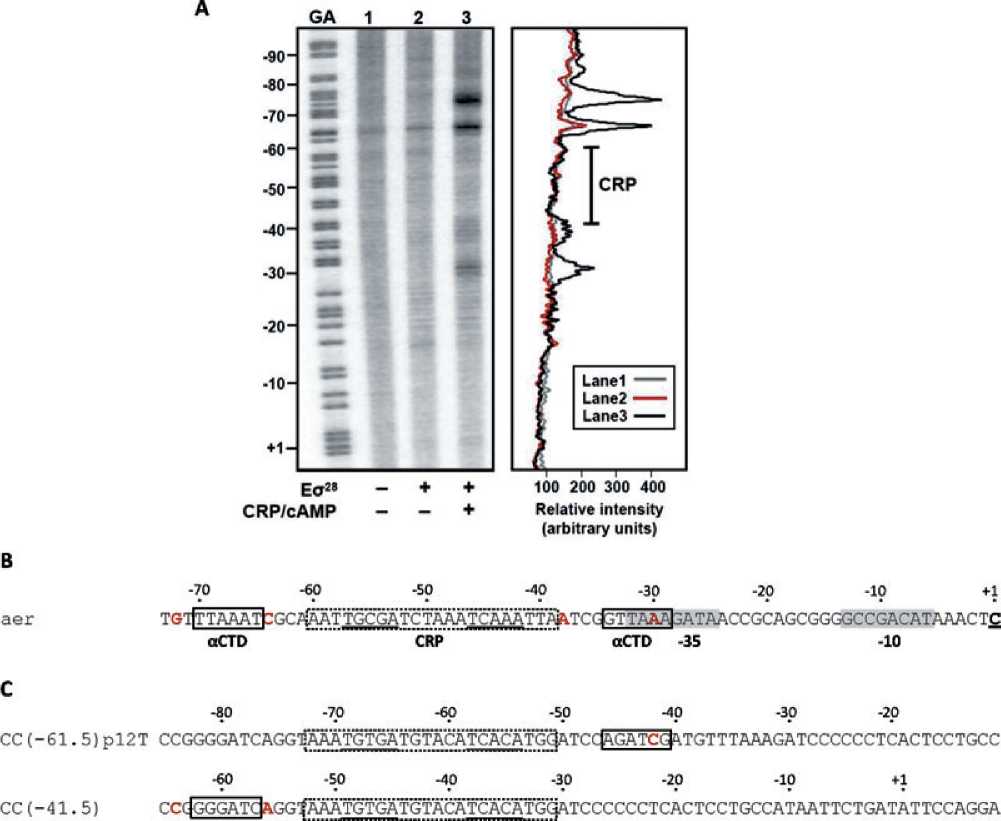
Fig. 5. Mapping the location of the RNA polymerase a C-terminal domains at the aer promoter.
A. The figure shows the results of FeBABE footprinting using the aer200 promoter fragment, and Es28 tagged with FeBABE at position 302 of
the a C-terminal domain. The PstI-HindIII promoter fragment, end-labelled on the template strand, was incubated in the presence or absence
of 200 nM FeBABE-tagged Es28 and 100 nM CRP/0.2 mM cAMP, as indicated. The left hand panel shows an autoradiograph of the 6%
polyacrylamide sequencing gel on which the reactions were run. The gel was calibrated using a Maxam-Gilbert ‘G + A’ sequencing reaction,
and is numbered with respect to the aer transcript start site. The right hand panel shows a plot of the relative intensity of bands down each
lane of the gel, with the position of the DNA site for CRP indicated.
B. Sequence of the aer promoter region, showing the proposed locations of aCTD binding. The -10 and -35 elements of the s28-dependent
promoter are shaded grey, the DNA site for CRP is denoted by the dashed box, and the locations of FeBABE-induced DNA cleavage are
highlighted in red. The sites where the aCTDs are proposed to contact the DNA (6 bp sequences centred 18-19 bp from the centre of the
DNA site for CRP (Benoff et al., 2002)) are indicated by the solid boxes. The sequence is numbered with respect to the aer transcript start
site.
C. Proposed locations of aCTD binding at the model Class I and Class II CRP-dependent promoters described by Lee et al. (2003). At each
promoter, the DNA site for CRP is denoted by a dashed box, and the locations of FeBABE-induced DNA cleavage are highlighted in red.
The sites where aCTD is proposed to contact the DNA are indicated by the solid boxes.
of CRP to specific targets in the tsr and trg regulatory
regions was also detected. Note that bioinformatic analy-
ses had predicted DNA sites for CRP upstream of both tsr
and trg (Robison et al., 1998). The trg promoter fragment
binds CRP with similar affinity to the aer fragment, while
CRP binding to the tsr fragment is much tighter. No clear
binding of CRP was found with the fliC/fliD, flgMN, flgKL,
motAB/cheAW or tar/tap/cheRBYZ fragments.
The action of CRP at the tsr and trg regulatory regions
was studied further. In the tsr regulatory region, the pre-
dicted CRP site is located 132.5 bp upstream of the s28-
dependent tsr promoter, so it is unlikely that CRP makes
direct contact with bound Es28. Indeed, no direct effect of
CRP on gene expression from the tsr regulatory region
could be detected (K. Hollands, unpubl. data). In contrast,
alignment of the DNA sequences of the trg and aer regu-
© 2009 The Authors
Journal compilation © 2009 Blackwell Publishing Ltd, Molecular Microbiology, 75, 1098-1111
More intriguing information
1. The name is absent2. The name is absent
3. An Estimated DSGE Model of the Indian Economy.
4. The name is absent
5. The Folklore of Sorting Algorithms
6. The Macroeconomic Determinants of Volatility in Precious Metals Markets
7. The name is absent
8. The Prohibition of the Proposed Springer-ProSiebenSat.1-Merger: How much Economics in German Merger Control?
9. CONSUMER ACCEPTANCE OF GENETICALLY MODIFIED FOODS
10. Structural Influences on Participation Rates: A Canada-U.S. Comparison